Keynote Speaker

Dr. Carlos Simmerling
Professor, Department of ChemistryAssociate Director, Laufer Center for Physical and Quantitative Biology, Stony Brook University, USA
Speech Title: Using Computer Simulations to Explore Vulnerabilities in the SARS-Cov-2 Spike Glycoprotein
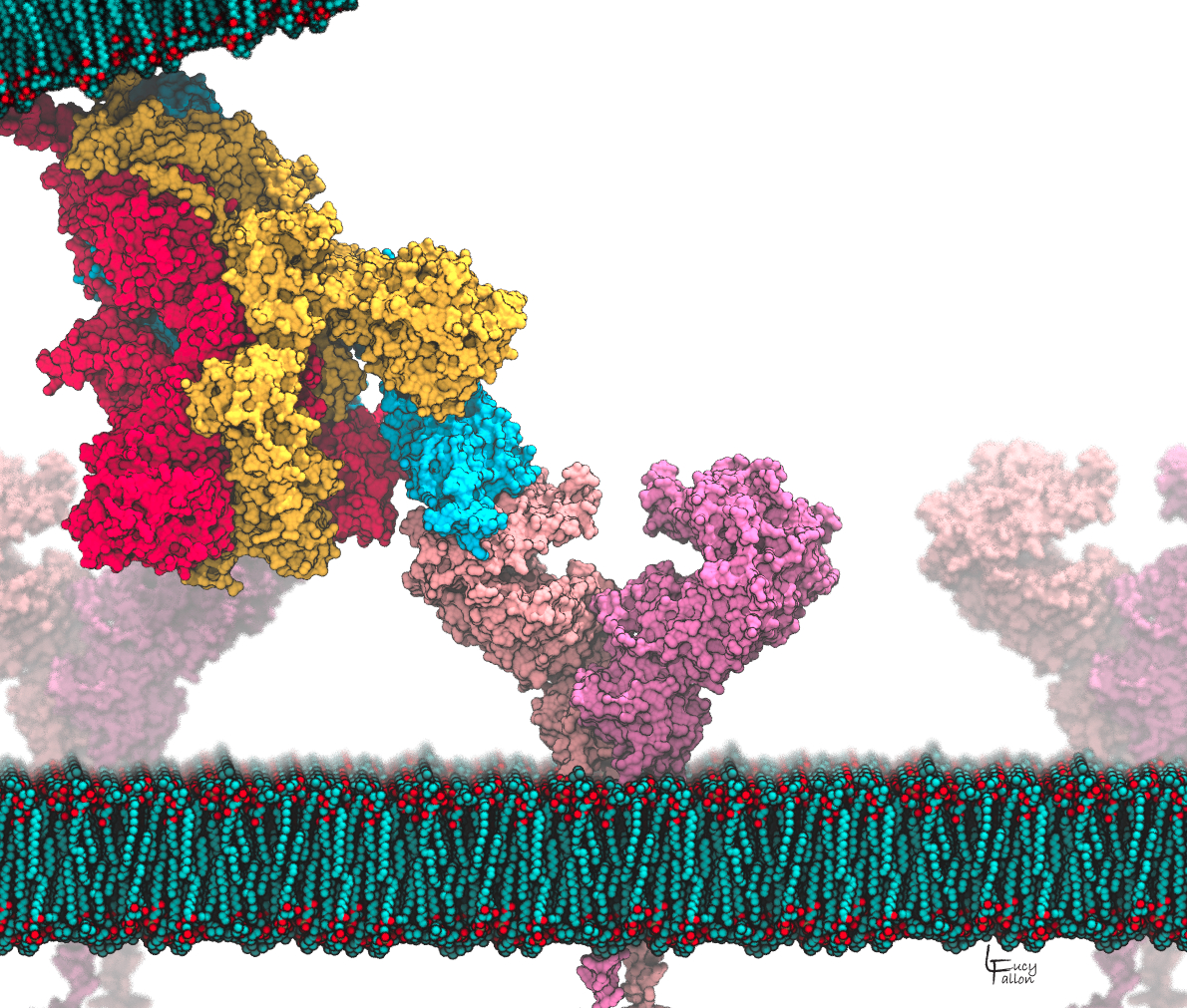
Abstract: Coronaviruses (CoVs) are so named due to the similarity of their appearance to a crown, with small protrusions of “spike” proteins covering their surface. The virus uses these spikes as molecular keys to unlock entry into and infect a host cell. Spike proteins are the key target of neutralizing antibodies, but development of immunity is slow. The spike has an extensive glycan shield that interferes with immune surveillance. Furthermore, antibodies tend to interact with the spike in the highly variable exposed regions, thus development of immunity to one CoV strain does not provide protection against another. We hypothesize that small molecule drugs could interact with the spike prior to immunity development, and block the conformational changes that are crucial to membrane fusion and infection. Modern drug discovery methods use structure to guide drug design, however these are hampered for COVID-19 because experimental structures are missing key regions for all coronavirus spikes, and none of the hypothesized membrane fusion steps have been directly observed. We are bridging this gap with computer models that can: complete the partial structures obtained from experiments, reveal the glycan shield absent in experimental models, provide mechanistic insight into viral evolution, and identify opportunities where drugs could block viral entry. Such drugs could have the potential to be broadly neutralizing of all CoVs, leading to effective treatments for COVID-19, as well as future pandemics caused by as-yet unknown CoVs.
Biography: Prof. Carlos Simmerling obtained his Bachelor’s degree (1991) and PhD (1994) in Chemistry at the University of Illinois at Chicago, performing early research on methods for computer modeling of biomolecules such as proteins. He went on to a post-doctoral fellowship in Pharmaceutical Chemistry at UCSF, where he became a lead developer of the Amber biomolecular simulation software that is used in thousands of research labs worldwide. In 1998 Prof. Simmerling joined the Chemistry department at Stony Brook University, where he is currently a Professor, and he became the Associate Director of SBU’s Laufer Center for Physical & Quantitative Biology. His research is funded by the USA National Institutes of Health, National Science Foundation, and Department of Energy. His work focuses on development of improved molecular simulation methods and models, and using these tools to study biomolecular recognition mechanisms. His articles on improving the physics underlying biomolecular modeling have been cited nearly 10,000 times. Prof. Simmerling is currently the Marsha Laufer Chair of Physical & Quantitative Biology at Stony Brook University and a Fellow of the American Chemical Society.
